- 1Univ Paris Est Creteil and Université Paris Cité, CNRS, LISA, F-94010 Créteil, France (clara.azemard@lisa.ipsl.fr)
- 2NASA Goddard Space Flight Center, 8800 Greenbelt Rd, Greenbelt, MD 20771, USA
- 3LATMOS, Institut Pierre Simon Laplace (IPSL), UMR CNRS 8190, UVSQ Université Paris-Saclay, Sorbonne Université, Guyancourt, France.
- 4CentraleSupelec, LGPM, 8-10 rue Joliot-Curie, 91190 Gif-sur-Yvette, France
- 5KapScience, LLC, Tewksbury, MA
- 6Danell Consulting, Inc., Winterville, NC 28590
- 7INAF-Arcetri Astrophysical Observatory, Florence, Italy
- 8RISE Research Institutes of Sweden, Stockholm, Sweden
- 9MPS, Gottingen, Germany
Introduction:
The ESA ExoMars rover mission mainly focuses on the search for potential life-relevant molecules (large, non-volatile organic or biological molecules that suggest current or ancient prebiotic activity) at the Mars surface and near-subsurface. The Mars Organic Molecule Analyzer (MOMA) aboard the Rosalind Franklin rover will be a key analytical tool to provide chemical (molecular) information from solid samples collected by the rover. The characterization of the organic content in the sample is the main purpose of the instrument. The MOMA instrument comprises a gas chromatograph coupled to a mass spectrometer (GC-MS) which offers the unique capability to analyze and identify a wide range of organic molecules, including species of interest for life and prebiotic chemistry. Here we present an evaluation of the performance of the instrument from the analysis of a full set of samples of interest for Mars, using the Engineering Test Unit of the GC and MS instruments.
Method:
The samples delivered to MOMA will be analyzed either by UV laser desorption/ionization ion trap mass spectrometry (LDI-ITMS) or pyrolysis gas chromatography ion trap mass spectrometry (pyr-GC-ITMS). Samples containing organic compounds previously detected by LDI-ITMS and/or pyr-GC-ITMS can undergo further analysis through reaction with chemical derivatization reagents before characterization. These reagents (MTBSTFA, DMF-DMA, or TMAH), stored inside sealed capsules, induce a chemical reaction with the sample (in particular with its organic compounds) enhancing the volatility of complex organic species. Due to the need to prevent organic contamination of the Flight Model (FM) and Spare Model (SM) of MOMA, it was not possible to perform extensive analytical tests on them. For that reason, a series of tests were performed by coupling the GC and the MS Engineering Test Units (ETU) which are very close replica of the FM on board the Exomars rover.
The ETU GC (developed in France by LISA and LATMOS laboratories) was mated to the ETU ion trap mass spectrometer (developed in the USA by Goddard Space Flight Center) in a flight-like configuration (Fig. 1) for the coupling campaign. The MOMA GC ETU includes a tank filled with the carrier gas (helium), four separate analytical modules including columns and thermal conductivity detectors (TCDs) and two thermal injection traps. A homemade oven designed and built at LISA is used to mimic MOMA FM ovens. During the pyrolysis of the sample or after its chemical derivatization with one of the three reagents, the volatile compounds are injected and pre-concentrated in one of the traps. The trap is then flash-heated with a backflush carrier gas flow to release the trapped chemical species as quickly as possible into one of the GC columns. The GC column separates the different molecules. They are then ionized by the electron impact ionization source in the MS chamber and analyzed by the linear ion trap mass spectrometer. During the ETU campaign, the derivatization reagents (MTBSTFA, DMF-DMA, and TMAH) are added inside the oven by a syringe mimicking the release of reagent from the MOMA capsules.
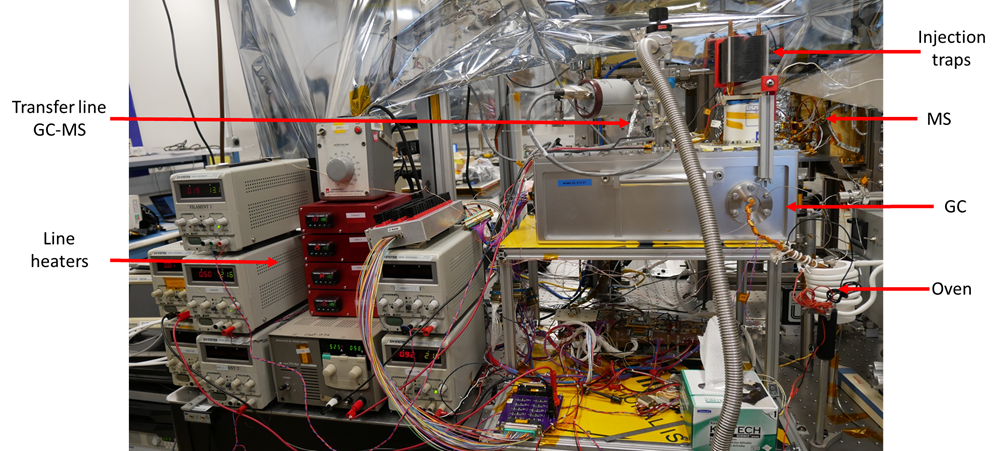
Figure 1: picture of the ETU gas chromatograph and the ETU MS ion trap.
Samples:
Several samples were studied during this campaign. Standard samples (standard gases, amino acids, carboxylic acids) were used to obtain the reference data. Then organic carbon-bearing natural and synthetic (mixing organic molecules and inorganic phases) Mars analog samples were analyzed, using both pyrolysis and derivatization as used in the MOMA instrument. These samples included JSC Mars-1, fragments of Murchison meteorite and synthetic samples composed of an organic-free mineral (vermiculite) doped with phenylalanine, phthalic acid and undecanoic acid. Also a synthetic “unknown” sample was used to run a blind analysis and to test the capability to identify a range of organic species under more realistic analytical conditions. This sample was used to simulate a day in the life of the instrument.
Results and conclusion:
The results described show the current state of end-to-end performance of the gas chromatography-mass spectrometry mode of operation. We show that from a technical point of view, the MOMA instrument works as expected. From a chemical perspective, we have shown that the four different types of MOMA analyses conducted here (pyrolysis and MTBSTFA, DMF-DMA, or TMAH derivatization) are efficient. Specific molecules have been detected (as expected) when using chemical standards. Analysis of the natural and synthetic samples reveals that a large number of molecules are released, and many species have been positively detected and identified (Fig. 2).
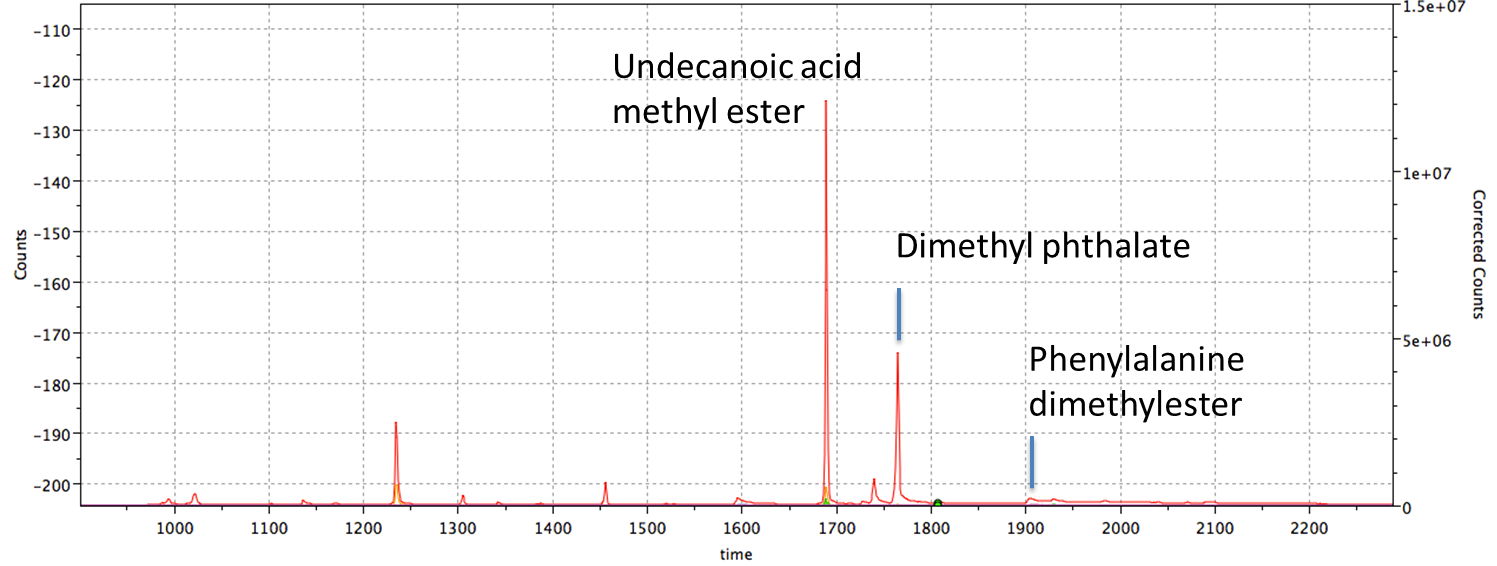
Figure 2 : Total ion chromatogram (TIC) for vermiculite doped with phenylalanine, phthalic acid and undecanoic acid after online DMF-DMA derivatization.
How to cite: Azémard, C., Stalport, F., Kaplan, D., Danell, R., Szopa, C., Buch, A., Chaouche, N., Freissinet, C., van Amerom, F., Grand, N., Zapf, P., Raulin, F., Guzman, M., Fornaro, T., Li, X., Grubisic, A., Siljeström, S., Cottin, H., Brinckerhoff, W. B., and Goesmann, F.: Analytical capabilities of the MOMA GC-MS instrument of the Exomars mission assessed from the analysis of a variety of samples with the Engineering Test Units (ETU), Europlanet Science Congress 2022, Granada, Spain, 18–23 Sep 2022, EPSC2022-1008, https://doi.org/10.5194/epsc2022-1008, 2022.

